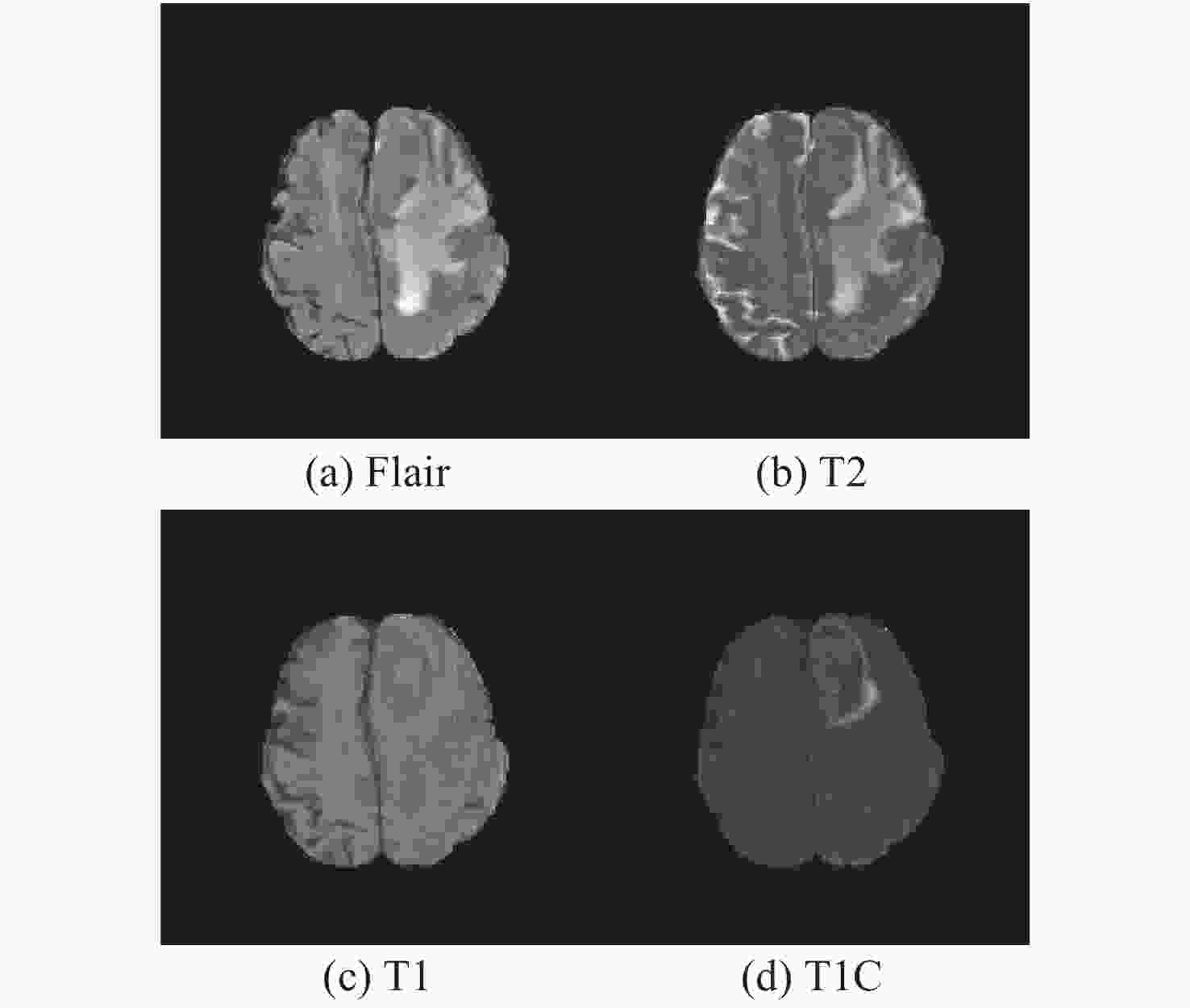
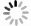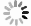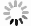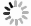Classification model based on fusion of multi-scale feature and channel feature for benign and malignant brain tumors
-
摘要:
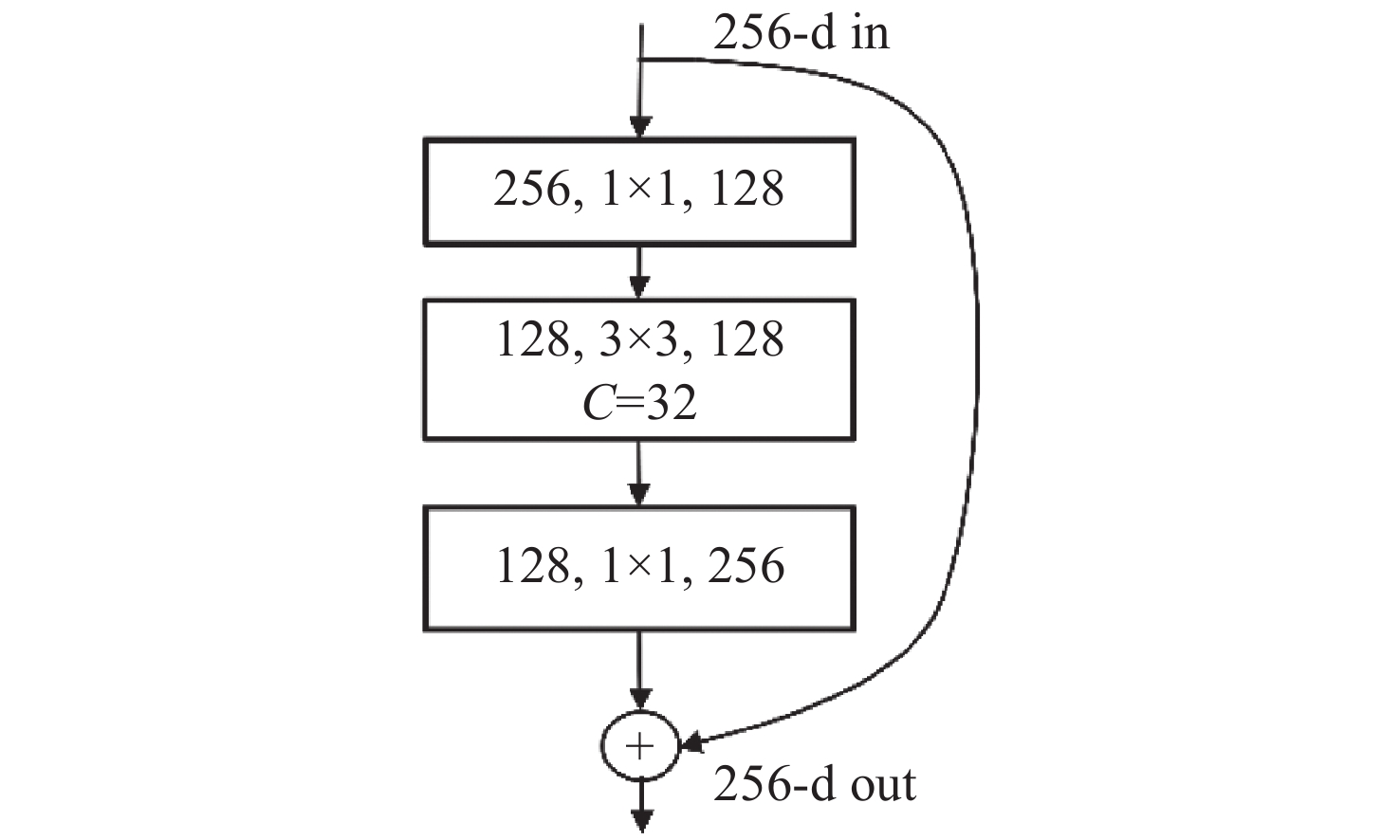
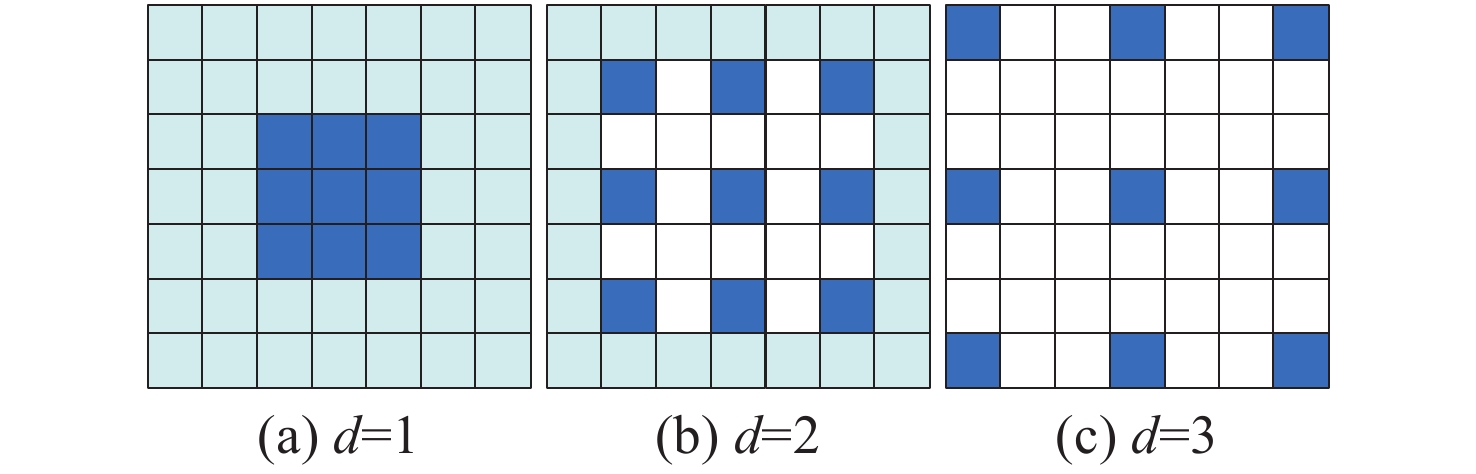
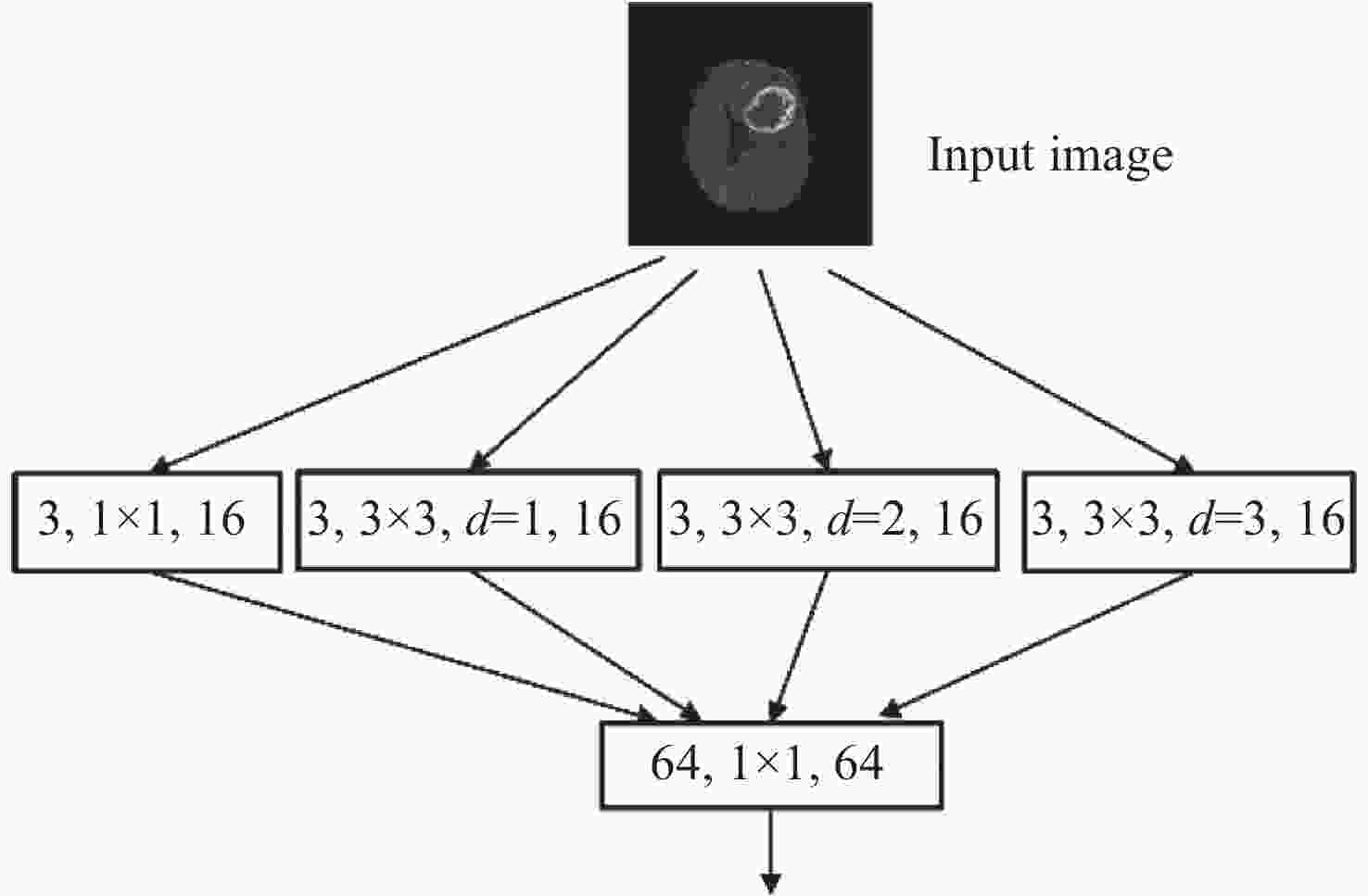
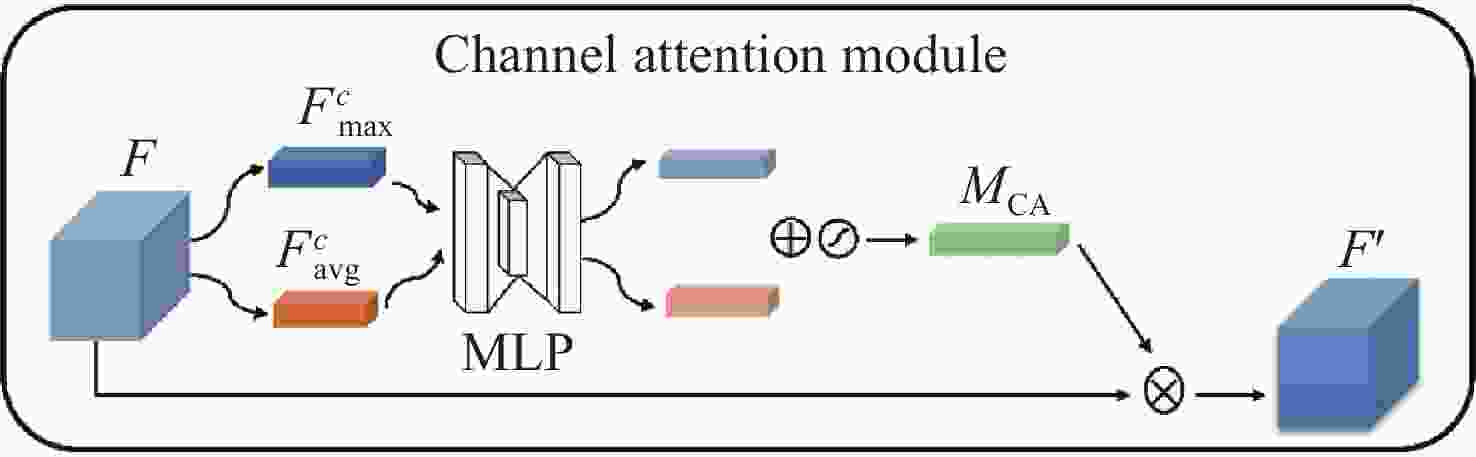
针对脑肿瘤良恶性分类过程复杂、分类准确率不高等问题,提出了一种基于多尺度特征与通道特征融合的分类模型。该模型以ResNeXt网络为主干网络,首先,将基于空洞卷积的多尺度特征提取模块代替第一层卷积层,利用膨胀率获取不同感受野的图像信息,将全局特征与局部显著特征相结合;其次,添加通道注意力机制模块,融合特征通道信息,提高对肿瘤区域的关注度,降低对冗余信息的关注度;最后,采用学习率的线性衰减策略、图像的标签平滑策略以及基于医学图像的迁移学习策略的组合优化提高模型的学习能力和泛化能力。在BraTS2017和BraTS2019数据集中进行实验,准确率分别达到98.11%和98.72%。与经典模型和其他先进方法相比,该分类模型能够有效地减少分类过程的复杂度,提高脑肿瘤良恶性分类的准确率。
Abstract:Aiming at the problems of complex and inaccurate classification of benign and malignant brain tumors, a classification model was proposed based on the fusion of multi-scale and channel features. The model used ResNeXt as the backbone network. First, the multi-scale feature extraction module based on dilated convolution was used to replace the first convolution layer, which can make full use of dilation rates to obtain the image information from different receptive fields, and combine the global features with significant subtle ones. Second, the channel attention mechanism module was added in the network to fuse the feature channel information in order to increase the attention to the tumor, and reduce the attention to redundant information. Finally, the combination optimization strategy, the MultiStepLR strategy of the learning rate, the label smoothing strategy and the transfer learning strategy on medical images were adopted to improve the learning and generalization abilities of the model. The experiments were carried out on BraTS2017 Dataset and BraTS2019 Dataset, and the classification accuracy were 98.11% and 98.72%, respectively. Compared with other advanced methods and classical models, the proposed classification model can effectively reduce the complexity of the classification process and improve the detection accuracy of benign and malignant brain tumors.
-
Key words:
- brain tumor /
- multi-scale feature /
- channel attention mechanism /
- deep learning
-
表 1 实验数据集分布
Table 1. Distribution of experimental datasets
数据集 肿瘤
类别数据分布 总数 训练集 测试集 BraTS2017
数据集HGG 840 210 1050 LGG 900 225 1125 BraTS2019
数据集HGG 1035 260 1295 LGG 915 225 1140 表 2 优化前BraTS2017数据集的分类结果评价表
Table 2. Evaluation of classification results on BraTS2017 before optimization
网络 ACC(%) SEN(%) SPE(%) PPV(%) NPV(%) ResNet 89.15±1.83 88.76±3.38 89.51±3.38 88.92±3.67 89.60±2.58 SENet 90.44±3.25 93.05±2.09 89.25±7.41 88.21±5.69 93.19±1.77 ResNeXt 90.34±1.14 89.23±3.36 92.53±2.18 91.85±1.96 90.22±2.57 MDCA-ResNeXt 93.19±0.35 93.05±1.67 93.33±1.69 92.91±1.54 93.54±1.36 表 3 优化前BraTS2019数据集的分类结果评价表
Table 3. Evaluation of classification results on BraTS2019 before optimization
网络 ACC(%) SEN(%) SPE(%) PPV(%) NPV(%) ResNet 91.83±2.73 93.31±2.12 90.13±4.59 91.71±3.65 92.10±2.51 SENet 91.91±2.42 90.54±5.63 91.74±4.83 93.00±3.76 93.25±2.54 ResNeXt 93.57±1.50 94.23±2.02 92.80±2.99 93.85±2.31 93.35±2.15 MDCA-ResNeXt 94.10±1.40 94.38±1.67 93.78±2.33 94.76±2.12 93.60±1.59 表 4 优化后BraTS2017数据集的分类结果评价表
Table 4. Evaluation of classification results on BraTS2017 after optimization
网络 ACC(%) SEN(%) SPE(%) PPV(%) NPV(%) Improved ResNet 96.87±1.49 96.76±0.71 96.98±2.90 96.84±2.97 96.98±0.64 Improved SENet 97.56±1.04 96.67±0.89 98.40±1.43 98.27±1.52 96.94±0.83 Improved ResNeXt 97.98±1.33 97.43±2.06 98.49±1.28 98.38±1.39 97.63±1.88 Improved MDCA-ResNeXt 98.11±0.41 97.43±0.26 98.76±0.91 98.66±0.97 97.63±0.22 表 5 优化后BraTS2019数据集的分类结果评价表
Table 5. Evaluation of classification results on BraTS2019 after optimization
网络 ACC(%) SEN(%) SPE(%) PPV(%) NPV(%) Improved ResNet 97.03±1.95 97.31±2.02 96.62±3.04 97.20±2.55 96.91±2.23 Improved SENet 96.69±0.88 94.38±1.89 98.74±0.93 98.62±1.01 94.99±1.57 Improved ResNeXt 97.98±0.57 97.69±0.47 98.31±0.73 98.53±0.64 97.36±0.54 Improved MDCA-ResNeXt 98.72±0.31 98.62±0.64 98.85±0.51 99.00±0.44 98.41±0.73 表 6 先进方法分类结果对比表
Table 6. Comparison of classification results of advanced methods
-
[1] BRAY F, FERLAY J, SOERJOMATARAM I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA:A Cancer Journal for Clinicians, 2018, 68(6): 394-424. doi: 10.3322/caac.21492 [2] BRAY F, FERLAY J, SOERJOMATARAM I, et al. Erratum: global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA:A Cancer Journal for Clinicians, 2020, 70(4): 313. [3] BAKAS S, AKBARI H, SOTIRAS A, et al. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features[J]. Scientific Data, 2017, 4: 170117. doi: 10.1038/sdata.2017.117 [4] MOHAN G, SUBASHINI M M. MRI based medical image analysis: survey on brain tumor grade classification[J]. Biomedical Signal Processing and Control, 2018, 39: 139-161. doi: 10.1016/j.bspc.2017.07.007 [5] KHARRAT A, HALIMA M B, AYED M B. MRI brain tumor classification using Support Vector Machines and meta-heuristic method[C]. IEEE 2015 15th International Conference on Intelligent Systems Design and Applications (ISDA), IEEE, 2015: 446-451. [6] 徐立, 白金牛, 李磊民. 基于脑部MR图像GMM特征决策分类的肿瘤诊断[J]. 控制工程,2017,24(8):1718-1722. doi: 10.14107/j.cnki.kzgc.150604XU L, BAI J N, LI L M. Diagnosis of tumor in brain MR images based on GMM features and decision tree classifier[J]. Control Engineering of China, 2017, 24(8): 1718-1722. (in Chinese) doi: 10.14107/j.cnki.kzgc.150604 [7] RAJU A R, SURESH P, RAO R R. Bayesian HCS-based multi-SVNN: a classification approach for brain tumor segmentation and classification using Bayesian fuzzy clustering[J]. Biocybernetics and Biomedical Engineering, 2018, 38(3): 646-660. doi: 10.1016/j.bbe.2018.05.001 [8] NARMATHA C, ELJACK S M, TUKA A A R M, et al. . A hybrid fuzzy brain-storm optimization algorithm for the classification of brain tumor MRI images[J]. Journal of Ambient Intelligence and Humanized Computing,doi: 10.1007/s12652-020-02470-5. [9] 黄乐弘, 曹立华, 李宁, 等. 深度学习的空间红外弱小目标状态感知方法[J]. 中国光学,2020,13(3):527-536.HUANG L H, CAO L H, LI N, et al. A state perception method for infrared dim and small targets with deep learning[J]. Chinese Optics, 2020, 13(3): 527-536. (in Chinese) [10] 郑江鹏, 余平, 赵萌, 等. 利用低信噪比小样本太赫兹光谱实现心肌淀粉样变检测[J]. 中国光学,2022,15(3):443-453. doi: 10.37188/CO.2021-0223ZHENG J P, YU P, ZHAO M, et al. Detection of myocardial amyloidosis by a small number of terahertz spectra with low signal-to-noise ratio[J]. Chinese Optics, 2022, 15(3): 443-453. (in Chinese) doi: 10.37188/CO.2021-0223 [11] 吴海滨, 魏喜盈, 王爱丽, 等. 八度卷积和双向门控循环单元结合的X光安检图像分类[J]. 中国光学,2020,13(5):1138-1146. doi: 10.37188/CO.2020-0073WU H B, WEI X Y, WANG A L, et al. X-ray security inspection images classification combined octave convolution and bidirectional GRU[J]. Chinese Optics, 2020, 13(5): 1138-1146. (in Chinese) doi: 10.37188/CO.2020-0073 [12] 李宇, 刘雪莹, 张洪群, 等. 基于卷积神经网络的光学遥感图像检[J]. 光学 精密工程,2018,26(1):200-207. doi: 10.3788/OPE.20182601.0200LI Y, LIU X Y, ZHANG H Q, et al. Optical remote sensing image retrieval based on convolutional neural networks[J]. Optics and Precision Engineering, 2018, 26(1): 200-207. (in Chinese) doi: 10.3788/OPE.20182601.0200 [13] 陈筱, 朱向冰, 吴昌凡, 等. 基于迁移学习与特征融合的眼底图像分类[J]. 光学 精密工程,2021,29(2):388-399. doi: 10.37188/OPE.20212902.0388CHEN X, ZHU X B, WU CH F, et al. Research on fundus image classification based on transfer learning and feature fusion[J]. Optics and Precision Engineering, 2021, 29(2): 388-399. (in Chinese) doi: 10.37188/OPE.20212902.0388 [14] GNANASEKARAN V S, JOYPAUL S, SUNDARAM P M, et al. Deep learning algorithm for breast masses classification in mammograms[J]. IET Image Processing, 2020, 14(12): 2860-2868. doi: 10.1049/iet-ipr.2020.0070 [15] SHARIF M I, LI J P, KHAN M A, et al. Active deep neural network features selection for segmentation and recognition of brain tumors using MRI images[J]. Pattern Recognition Letters, 2020, 129: 181-189. doi: 10.1016/j.patrec.2019.11.019 [16] KHAN M A, ASHRAF I, ALHAISONI M, et al. Multimodal brain tumor classification using deep learning and robust feature selection: a machine learning application for radiologists[J]. Diagnostics, 2020, 10(8): 565. doi: 10.3390/diagnostics10080565 [17] REHMAN A, KHAN M A, SABA T, et al. Microscopic brain tumor detection and classification using 3D CNN and feature selection architecture[J]. Microscopy Research and Technique, 2021, 84(1): 133-149. doi: 10.1002/jemt.23597 [18] SEETHA J, RAJA S S. Brain tumor classification using Convolutional Neural Networks[J]. Biomedical and Pharmacology Journal, 2018, 11(3): 1457-1461. doi: 10.13005/bpj/1511 [19] 赵尚义, 王远军. 基于3D CNN的脑胶质瘤分类算法[J]. 光学技术,2019,45(6):749-755.ZHAO SH Y, WANG Y J. Brain glioma classification algorithm based on 3D CNN[J]. Optical Technique, 2019, 45(6): 749-755. (in Chinese) [20] XIE S N, GIRSHICK R, DOLLÁR P, et al. . Aggregated residual transformations for deep neural networks[C]. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), IEEE, 2017: 5987-5995. [21] SZEGEDY C, VANHOUCKE V, IOFFE S, et al. . Rethinking the inception architecture for computer vision[C]. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), IEEE, 2016: 2818-2826. [22] HE K M, ZHANG X Y, REN SH Q, et al. . Deep residual learning for image recognition[C]. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), IEEE, 2016: 770-778. [23] YU F, KOLTUN V. Multi-scale context aggregation by dilated convolutions[C]. 4th International Conference on Learning Representations, 2016: https://doi.org/10.48550/arXiv.1511.07122. [24] WOO S, PARK J, LEE J Y, et al. . CBAM: convolutional block attention module[C]. Proceedings of the 15th European Conference on Computer Vision, Springer, 2018: 3-19. [25] JIANG L Q, NING CH Y, LI J Y. Glioma classification framework based on SE-ResNeXt network and its optimization[J]. IET Image Processing, 2022, 16(2): 596-605. doi: 10.1049/ipr2.12374 [26] CHENG J, HUANG W, CAO SH L, et al. Enhanced performance of brain tumor classification via tumor region augmentation and partition[J]. PLoS One, 2015, 10(12): e0140381. [27] VOVK U, PERNUS F, LIKAR B. A review of methods for correction of intensity inhomogeneity in MRI[J]. IEEE Transactions on Medical Imaging, 2007, 26(3): 405-421. doi: 10.1109/TMI.2006.891486 [28] TUSTISON N J, AVANTS B B, COOK P A, et al. N4ITK: improved N3 bias correction[J]. IEEE Transactions on Medical Imaging, 2010, 29(6): 1310-1320. doi: 10.1109/TMI.2010.2046908 [29] ZHONG ZH, ZHENG L, KANG G L, et al. . Random erasing data augmentation[C]. Proceedings of the 34th AAAI Conference on Artificial Intelligence, AAAI, 2020: 13001-13008. [30] HU J, SHEN L, SUN G. Squeeze-and-excitation networks[C]. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), IEEE, 2018: 7132-7141. -





 下载:
下载: