Advances in multi-dimensional single molecule imaging
-
摘要:
单分子成像方法被广泛应用于亚细胞结构的三维空间定位。点扩散函数是分析单分子信息的重要窗口,除了能反映空间坐标外还蕴含着丰富的额外信息。本文介绍了从点扩散函数中解析空间位置、荧光波长、偶极子朝向及干涉相位等多维度单分子成像研究进展,简要地概括了目前主流定位方法,并对该技术的发展方向进行了展望。
Abstract:Single-molecule imaging is widely used for the reconstruction of three-dimensional subcellular structures. The point spread function is an important window to analyze the information of a single molecule. Besides 3D coordinates, it also contains abundant additional information. In this paper, we reviewed the recent progress of multi-dimensional single-molecule imaging, including spatial location, fluorescence wavelength, dipole orientation, interference phase, etc. We also briefly introduced the latest methods for molecule localization and proposed the further directions for its research.
-
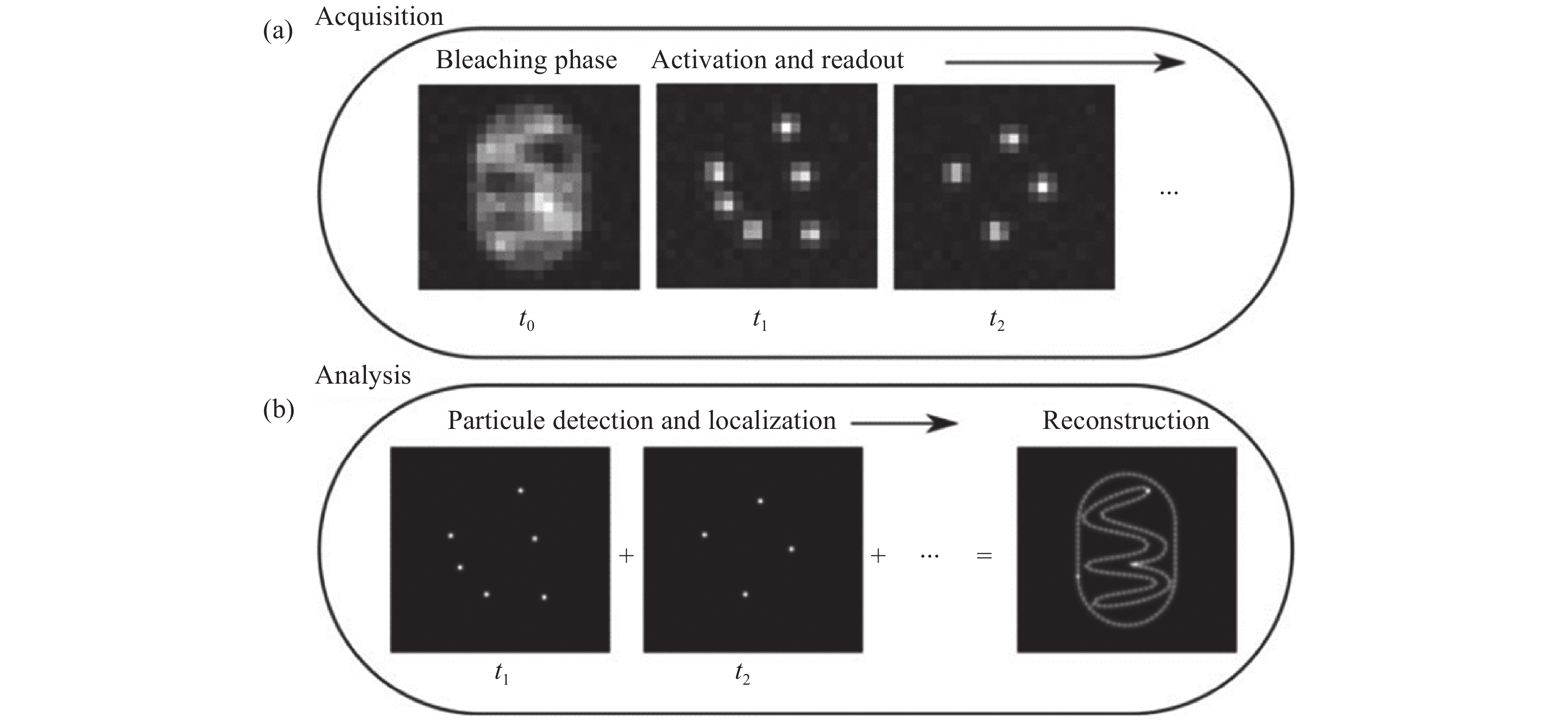
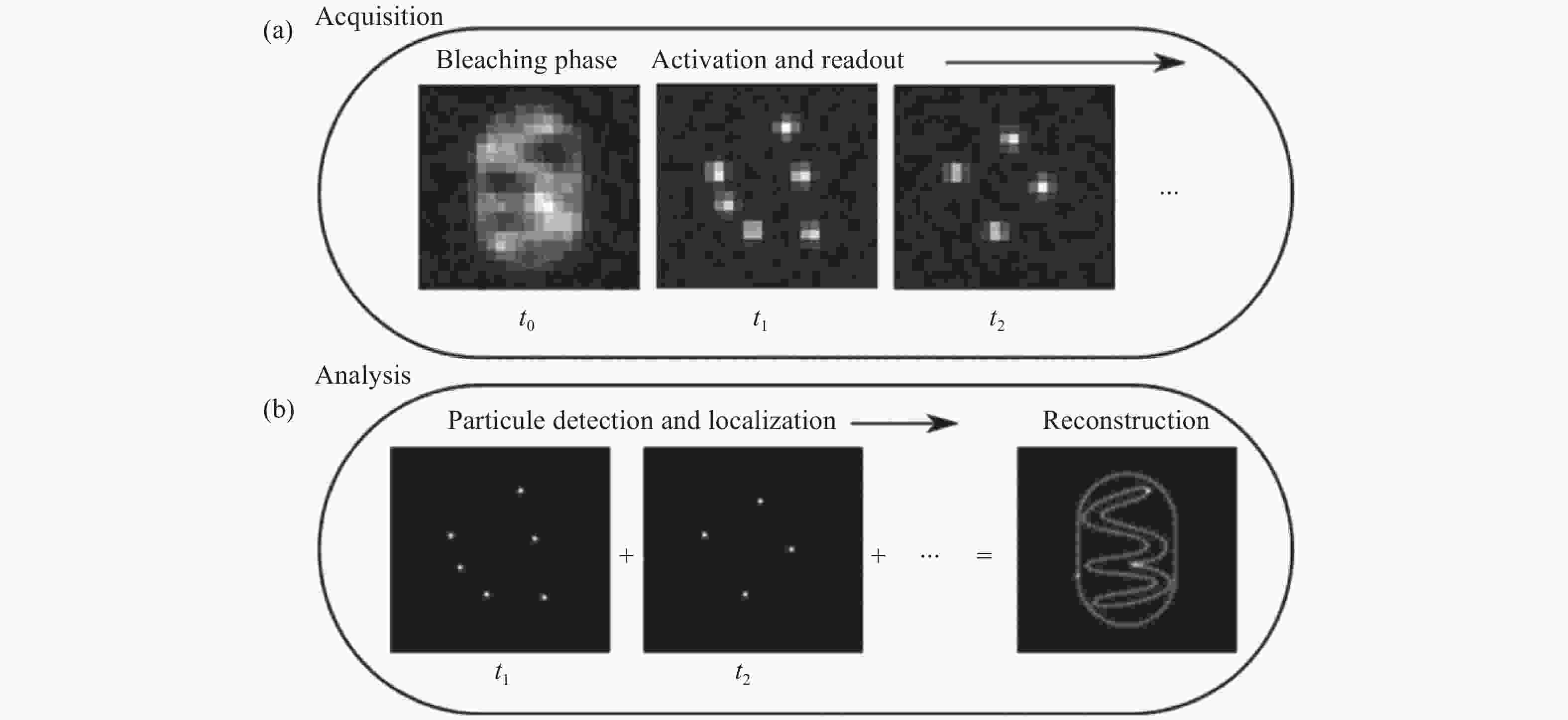
图 1 单分子二维定位[21]。(a)在采集步骤中,将会获取稀疏分布的单分子闪烁图像;(b)分析步骤中,从单帧图像中准确定位的单分子二维位置,以及所有单分子点的合成图像。
Figure 1. Two-dimensional localization of a single molecule [21]. (a) In the acquisition step, sparsely distributed single molecule images are recorded; (b) in the analysis step, the two-dimensional coordinates of the single molecules are precisely localized in each frame and then accumulated to reconstruct the super-resolution image
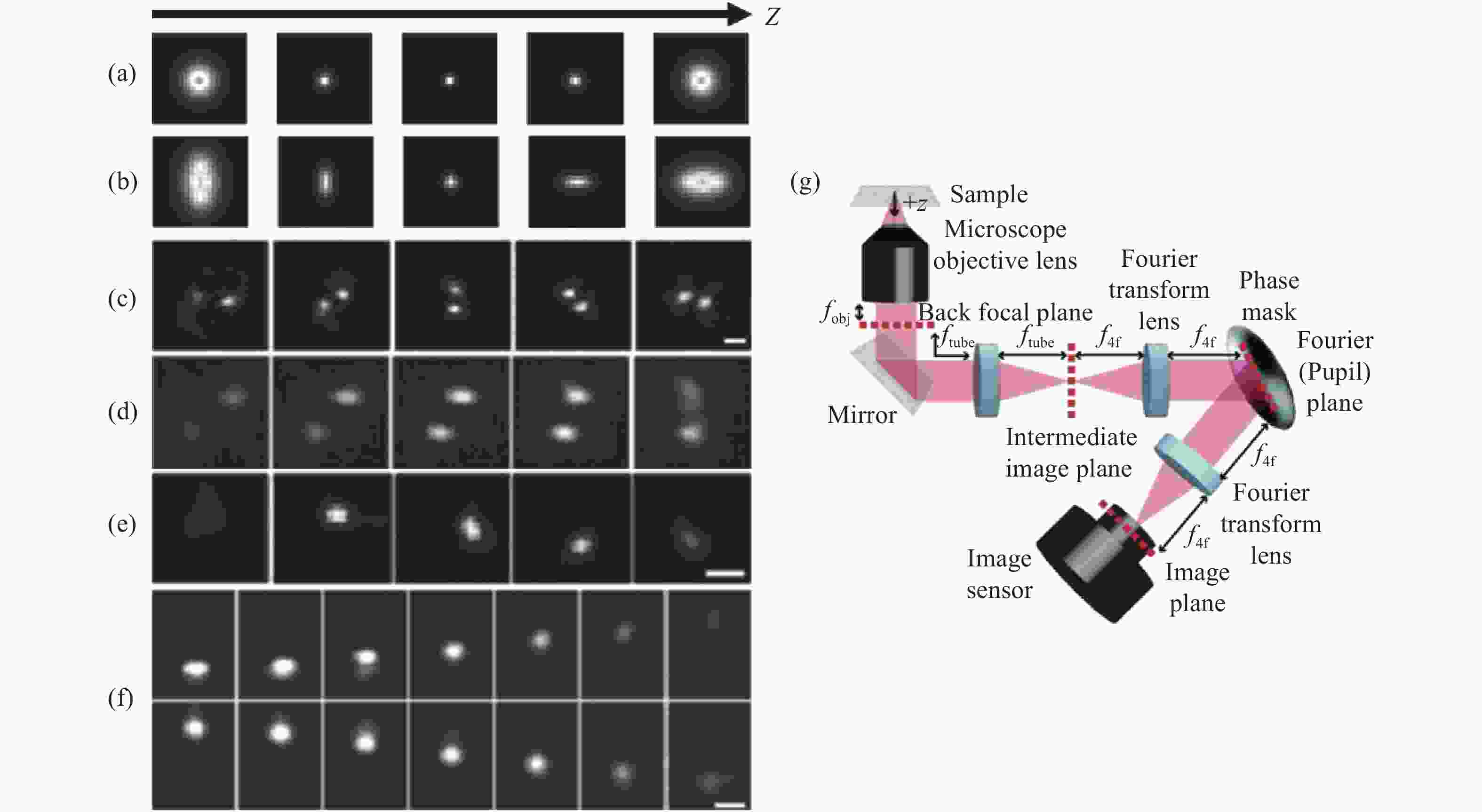
图 2 各PSF在不同轴向位置的变化及在SLM调制下的光路布局。(a)标准PSF;(b)散光PSF;(c)双螺旋PSF[27-28];(d)相位斜坡PSF[29];(e)螺旋PSF[30];(f)自弯曲PSF[33];(g) SLM调制下的光路布局
Figure 2. Changes of each PSF at different axial positions and optical path layout for SLM modulation. (a) The standard PSF; (b) astigmatism PSF; (c) double helix PSF[27-28]; (d) phase ramp PSF[29]; (e) spiral PSF[30]; (f) self-bending PSF[33]; (g) optical path layout for SLM modulation
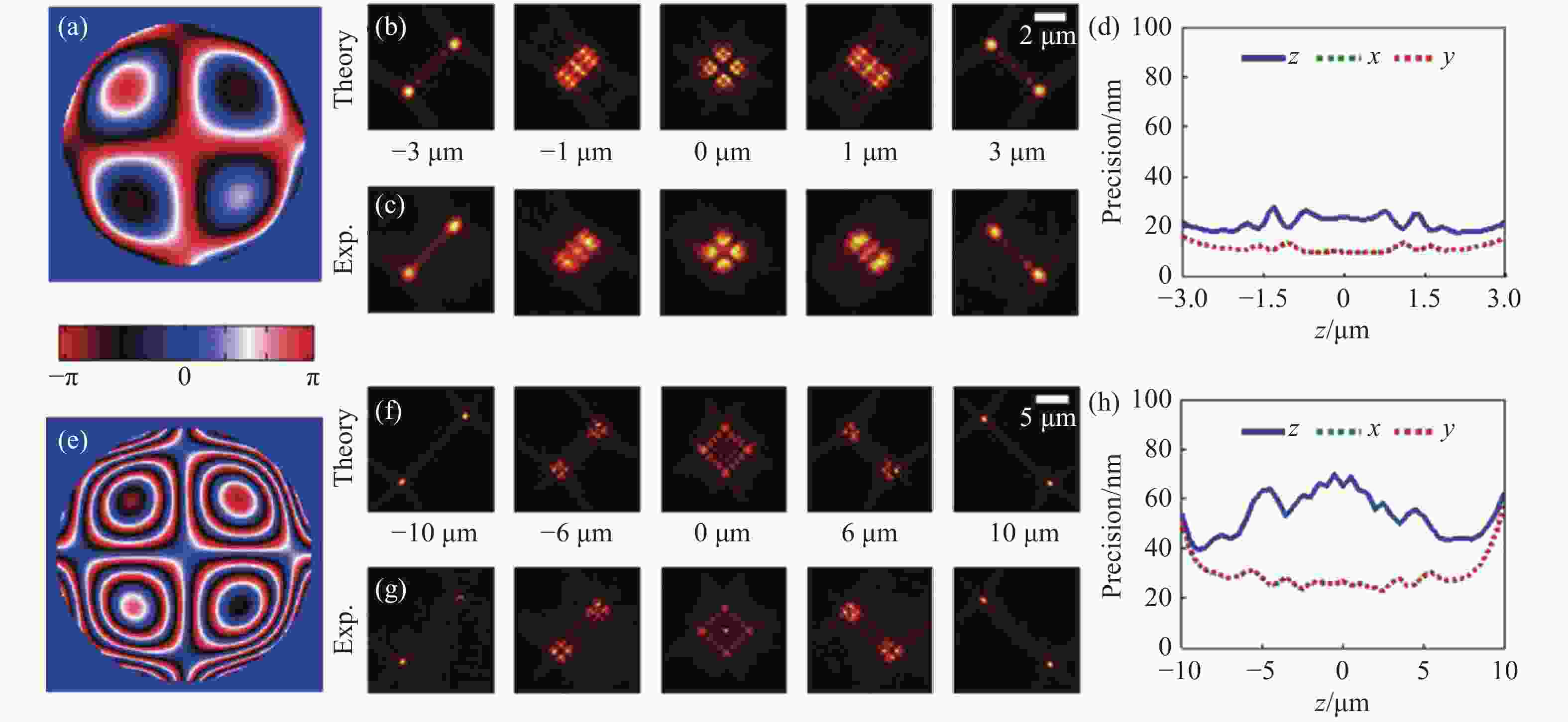
图 3 不同景深优化下的Tetrapod PSF[37]。6 µm优化景深下的光瞳函数(a),理论PSF(b),实验PSF(c),定位精度(d)。(e)~(h)与(a)~(d)相同,但是为10 μm优化景深下的Tetrapod PSF。
Figure 3. Tetrapod PSF optimized at different depths of field[37]. (a) The pupil function, (b) theoretical PSF, (c) experimental PSF, (d) localizing accuracy of Tetrapod PSF optimized for 6 µm depth of field. (e)~(h) The same as (a)~(d), but for Tetrapod PSF optimized for 10 µm depth of field.
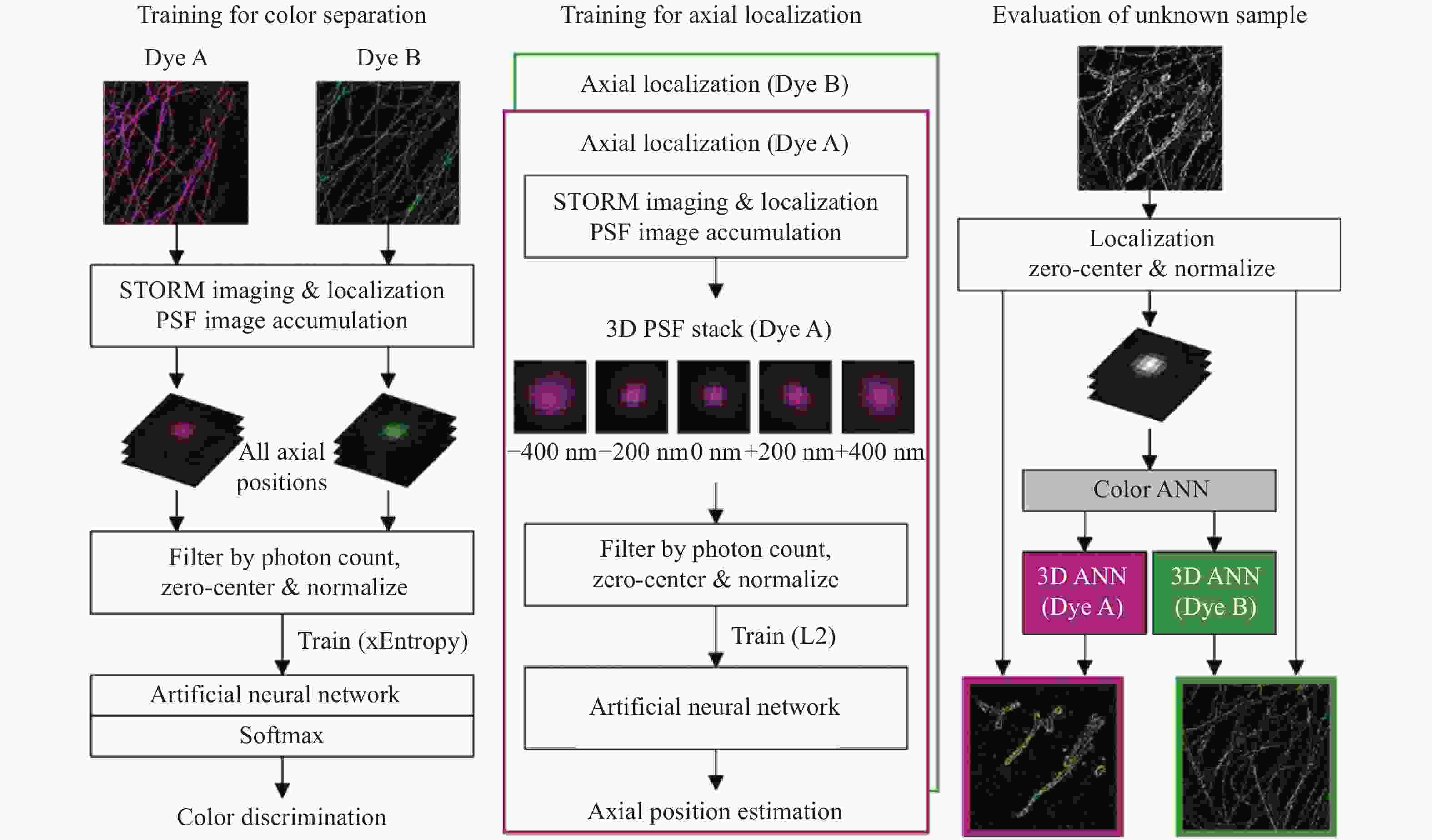
图 4 同时测量单分子的发射波长与三维位置 [48] 。(a)光路设计图,SLM放置在后焦面上;(b)弯曲光栅的光瞳函数;(c)3种波长在不同位置下的PSF分布,波长越长2个旁瓣的距离越远
Figure 4. Simultaneous measurement of emission wavelength and 3D position of single molecules[48]. (a) Optical path design with an SLM placed on the back focal plane; (b) pupil function of curved grating; (c) PSF distributions of three different wavelengths at different localizations. The longer the wavelength, the farther the distance between the two side lobes
图 6 偶极子方向引起的定位偏差[53]。(a) 转动角、极角、方位角分别为15°、45°、0°时单分子点的PSF xz切面(左图),xy切面(右图),以及其定位偏差;(b)极角、方位角与(a)相同的情况下转动角为60°的PSF;(c)和(d)分别为不同转动角,极角产生的横向偏移值;(e)转动角、极角、方位角在偶极子中的物理意义
Figure 6. Localization deviation caused by the dipole’s direction[53]. (a) PSF xz section (left) and xy section (right) of single molecule with a rotation angle, polar angle and azimuth angle of 15°, 45° and 0° respectively, and corresponding localization deviations; (b) PSF with the same polar angle and azimuth angle as (a) and rotation angle of 60°. (c) and (d) are the lateral offsets generated by different rotation angles and polar angles, respectively; (e) physical meaning of rotation angle, polar angle and azimuth angle of the dipole
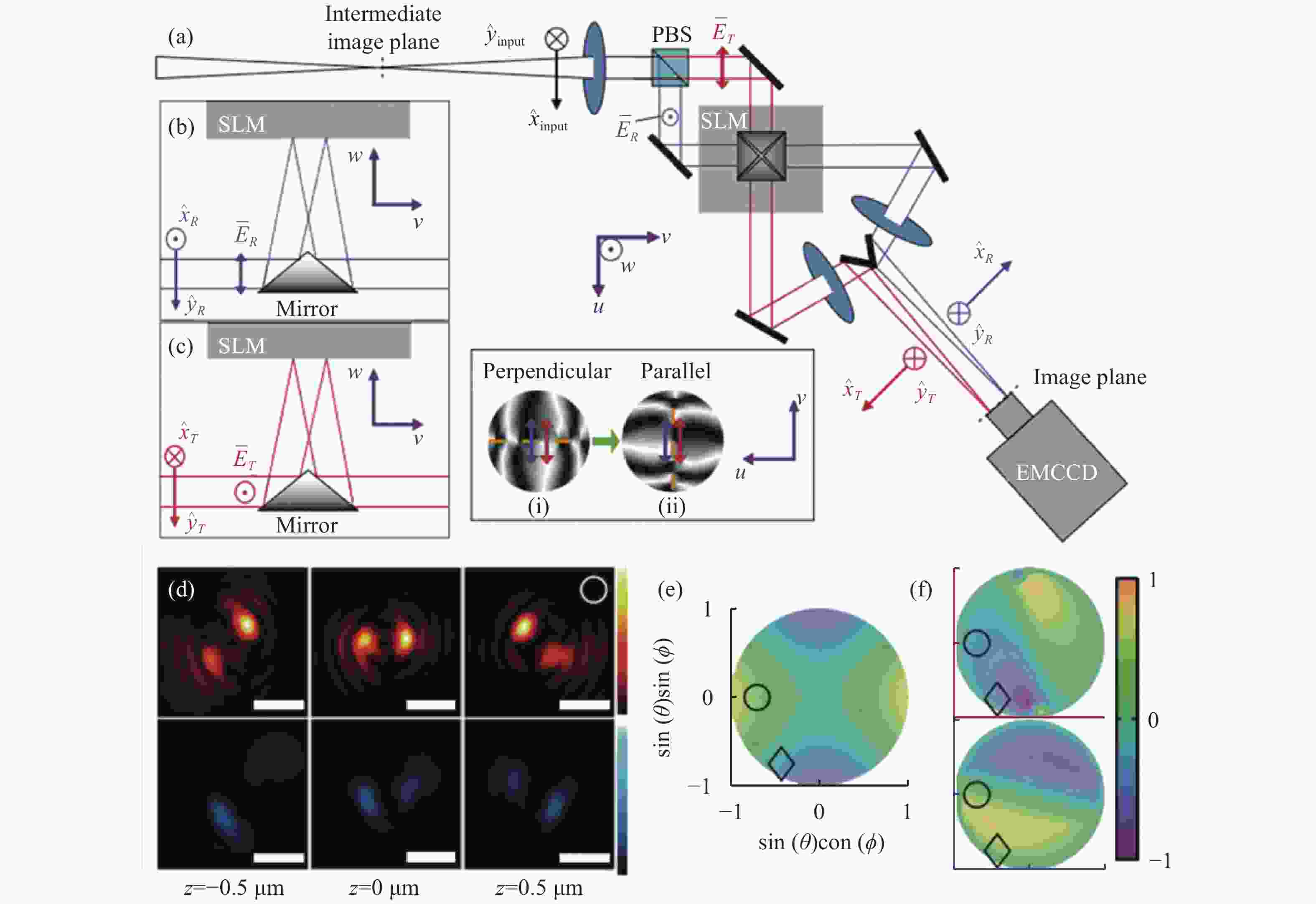
图 7 基于双螺旋PSF的偶极子方向定位方法[58]。(a)光路布局;(b)和(c)分别为两个偏振方向的成像通道,它们的光瞳函数分别为(i)、(ii);(d)上下图分别为水平和垂直通道的PSF;(e)和(f)分别为LA、LD指标,只考虑LA指标时会出现4种可能的朝向结果。红色和蓝色分别代表透射通道和反射通道的LD指标
Figure 7. Dipole orientation localization method based on double helix PSF[58] . (a) Optical path layout; (b) and (c) are imaging channels in two polarization directions, respectively, and their pupil functions are (i) and (ii) respectively; (d) the upper and lower figures are PSF of horizontal and vertical channels, respectively; (e) and (f) are LA and LD indicators respectively. There are four possible orientations when only the LA indicator is considered. Red and blue represent LD indexes of transmission channel and reflection channel respectively
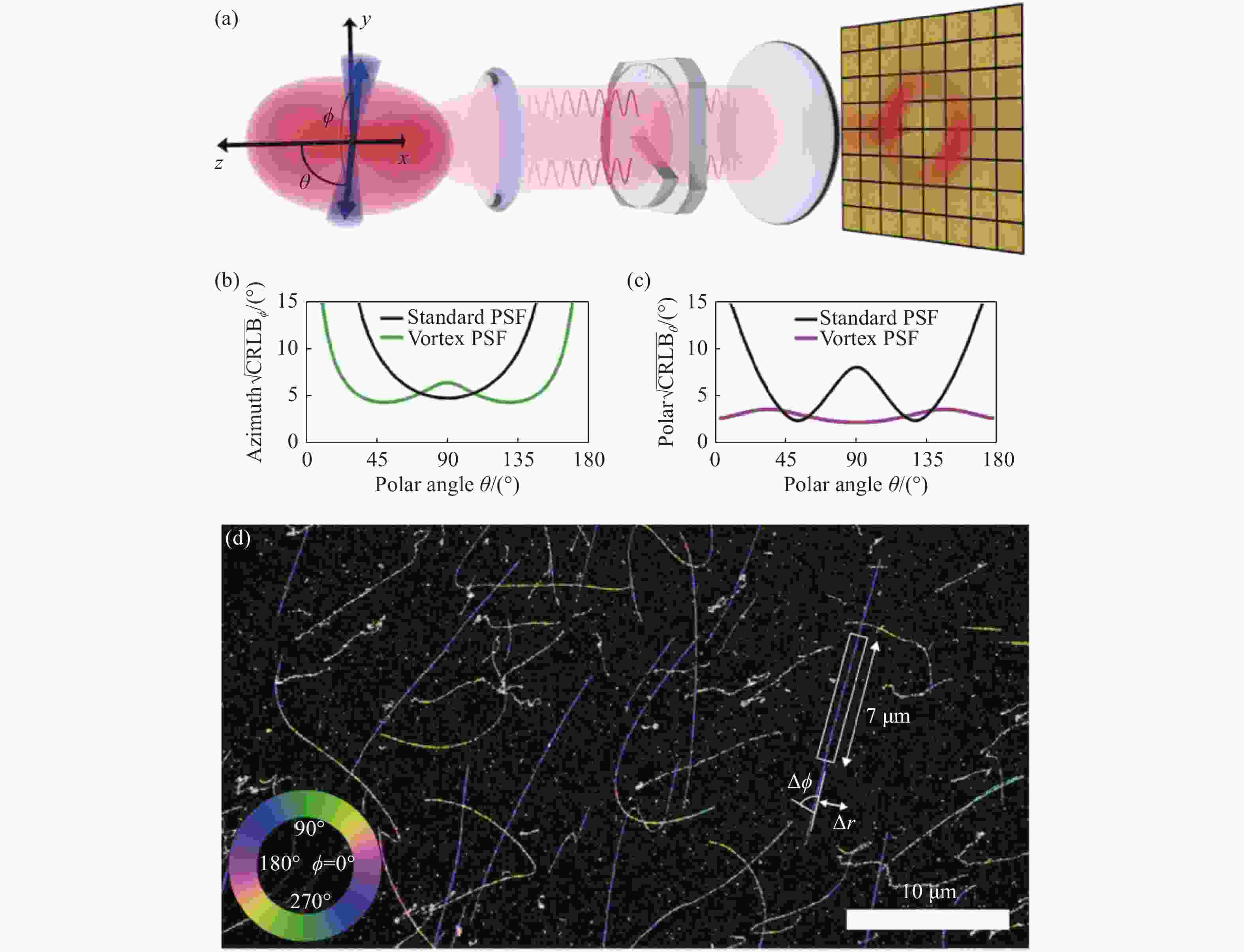
图 8 基于vortex PSF的朝向与三维位置同时定位[60]。(a)Vortex PSF光路;在4000个光子10个背景光子的单分子图像中,(b)方位角φ和(c)极角θ的CRLB;(d)λ-DNA的二维位置及方位角,伪色代表该点方位角,大小如左下角
Figure 8. Simultaneous localization of the single-molecule orientation and three-dimensional location based on vortex PSF[60] . (a) Vortex PSF’s optical path; CRLB of azimuth angle (b) and polar angle (c) in single molecule imaging with 4000 photons and 10 background photons; (d) 2D position and azimuthal angle of the λ -DNA. The false color represents the azimuthal angle, as shown in the lower left corner
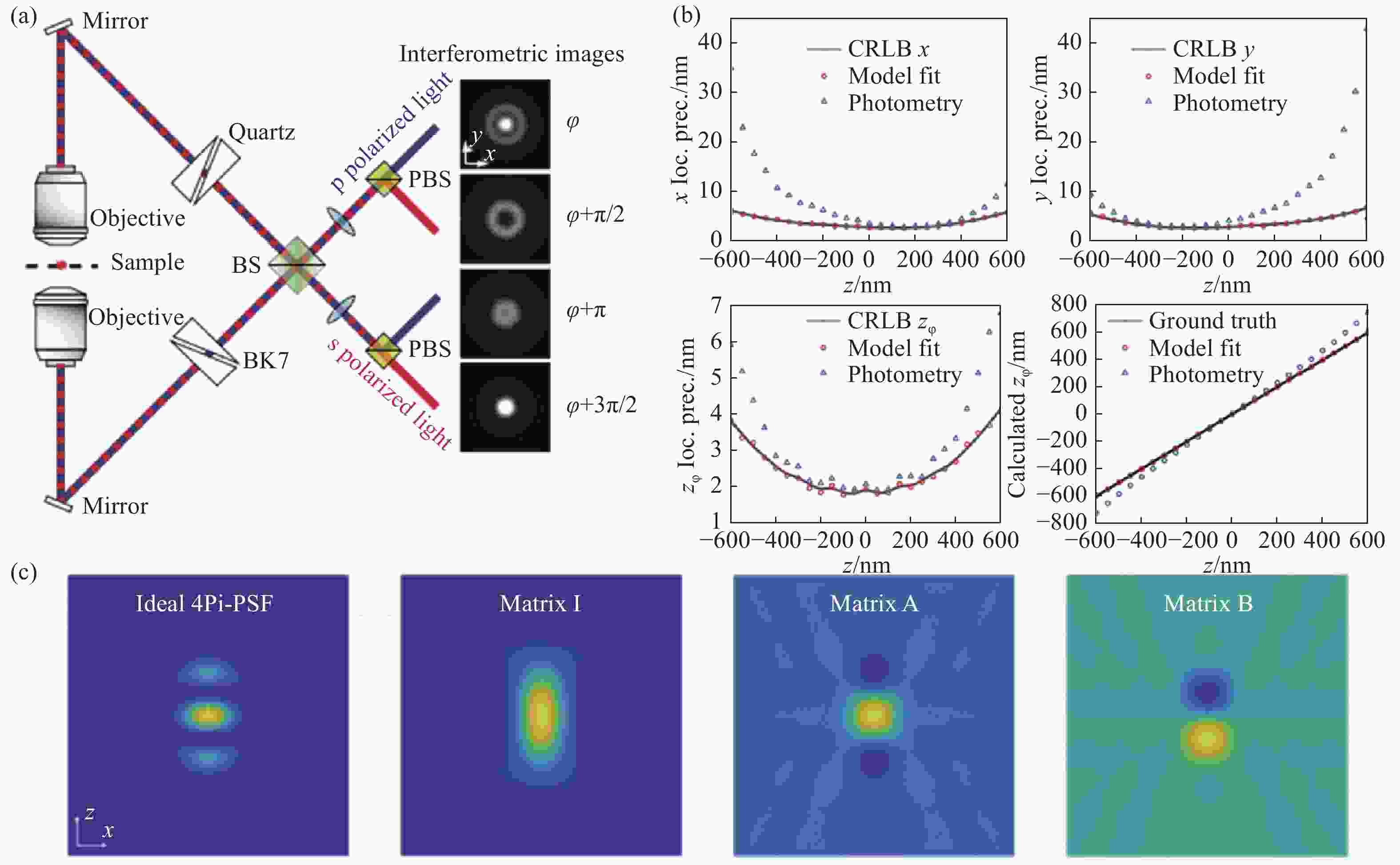
图 9 IAB模型分解的4Pi-PSF[65]。(a)四通道4Pi-SMLM光路结构以及各通道的PSF;(b)每个物镜接收2000光子以及20个背景光子下,通过光度法和IAB模型拟合得到的各维度定位精度;(c)理想的4Pi-PSF与分解出来的IAB矩阵
Figure 9. 4Pi-PSF decomposed by IAB model[65]. (a) The optical path layout of 4-channel 4Pi-SMLM and the PSFs of each channel; (b) localization accuracy of each dimension obtained by photometric method and IAB model fitting for single molecule of 2,000 photons collected by each objective lens; (c) ideal 4Pi-PSF and decomposed IAB matrix
-
[1] GUSTAFSSON M G L. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy[J]. Journal of Microscopy, 2000, 198(2): 82-87. doi: 10.1046/j.1365-2818.2000.00710.x [2] GUSTAFSSON M G L. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(37): 13081-13086. doi: 10.1073/pnas.0406877102 [3] BLOM H, WIDENGREN J. Stimulated emission depletion microscopy[J]. Chemical Reviews, 2017, 117(11): 7377-7427. doi: 10.1021/acs.chemrev.6b00653 [4] HELL S W, WICHMANN J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy[J]. Optics Letters, 1994, 19(11): 780-782. doi: 10.1364/OL.19.000780 [5] KLAR T A, JAKOBS S, DYBA M, et al. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission[J]. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(15): 8206-8210. doi: 10.1073/pnas.97.15.8206 [6] RUST M J, BATES M, ZHUANG X W. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM)[J]. Nature Methods, 2006, 3(10): 793-796. doi: 10.1038/nmeth929 [7] HEILEMANN M, VAN DE LINDE S, SCHÜTTPELZ M, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes[J]. Angewandte Chemie International Edition, 2008, 47(33): 6172-6176. doi: 10.1002/anie.200802376 [8] ZHUANG X W. Nano-imaging with STORM[J]. Nature Photonics, 2009, 3(7): 365-367. doi: 10.1038/nphoton.2009.101 [9] BETZIG E, PATTERSON G H, SOUGRAT R, et al. Imaging intracellular fluorescent proteins at nanometer resolution[J]. Science, 2006, 313(5793): 1642-1645. doi: 10.1126/science.1127344 [10] SHTENGEL G, GALBRAITH J A, GALBRAITH C G, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(9): 3125-3130. doi: 10.1073/pnas.0813131106 [11] SHROFF H, WHITE H, BETZIG E. Photoactivated localization microscopy (PALM) of adhesion complexes[J]. Current Protocols in Cell Biology, 2013, 58(1): 4.21.1-24.21.28. [12] LIU Y J, LU Y Q, YANG X S, et al. Amplified stimulated emission in upconversion nanoparticles for super-resolution nanoscopy[J]. Nature, 2017, 543(7644): 229-233. doi: 10.1038/nature21366 [13] ZHAN Q Q, LIU H CH, WANG B J, et al. Achieving high-efficiency emission depletion nanoscopy by employing cross relaxation in upconversion nanoparticles[J]. Nature Communications, 2017, 8(1): 1058. doi: 10.1038/s41467-017-01141-y [14] LIANG L L, FENG Z W, ZHANG Q M, et al. Continuous-wave near-infrared stimulated-emission depletion microscopy using downshifting lanthanide nanoparticles[J]. Nature Nanotechnology, 2021, 16(9): 975-980. doi: 10.1038/s41565-021-00927-y [15] SCHNITZBAUER J, STRAUSS M T, SCHLICHTHAERLE T, et al. Super-resolution microscopy with DNA-PAINT[J]. Nature Protocols, 2017, 12(6): 1198-1228. doi: 10.1038/nprot.2017.024 [16] SCHUEDER F, LARA-GUTIéRREZ J, BELIVEAU B J, et al. Multiplexed 3D super-resolution imaging of whole cells using spinning disk confocal microscopy and DNA-PAINT[J]. Nature Communications, 2017, 8(1): 2090. doi: 10.1038/s41467-017-02028-8 [17] JIA H, YANG J K, LI X J. Minimum variance unbiased subpixel centroid estimation of point image limited by photon shot noise[J]. Journal of the Optical Society of America A, 2010, 27(9): 2038-2045. doi: 10.1364/JOSAA.27.002038 [18] STALLINGA S, RIEGER B. Accuracy of the gaussian point spread function model in 2D localization microscopy[J]. Optics Express, 2010, 18(24): 24461-24476. doi: 10.1364/OE.18.024461 [19] SMALL A, STAHLHEBER S. Fluorophore localization algorithms for super-resolution microscopy[J]. Nature Methods, 2014, 11(3): 267-279. doi: 10.1038/nmeth.2844 [20] PATTERSON G, DAVIDSON M, MANLEY S, et al. Superresolution imaging using single-molecule localization[J]. Annual Review of Physical Chemistry, 2010, 61: 345-367. doi: 10.1146/annurev.physchem.012809.103444 [21] HERBERT S, SOARES H, ZIMMER C, et al. Single-molecule localization super-resolution microscopy: deeper and faster[J]. Microscopy and Microanalysis, 2012, 18(6): 1419-1429. doi: 10.1017/S1431927612013347 [22] HUANG B, WANG W Q, BATES M, et al. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy[J]. Science, 2008, 319(5864): 810-813. doi: 10.1126/science.1153529 [23] HOLTZER L, MECKEL T, SCHMIDT T. Nanometric three-dimensional tracking of individual quantum dots in cells[J]. Applied Physics Letters, 2007, 90(5): 053902. doi: 10.1063/1.2437066 [24] FU SH, LI M F, ZHOU L L, et al. Deformable mirror based optimal PSF engineering for 3D super-resolution imaging[J]. Optics Letters, 2022, 47(12): 3031-3034. doi: 10.1364/OL.460949 [25] PIESTUN R, SCHECHNER Y Y, SHAMIR J. Propagation-invariant wave fields with finite energy[J]. Journal of the Optical Society of America A, 2000, 17(2): 294-303. doi: 10.1364/JOSAA.17.000294 [26] GREENGARD A, SCHECHNER Y Y, PIESTUN R. Depth from diffracted rotation[J]. Optics Letters, 2006, 31(2): 181-183. doi: 10.1364/OL.31.000181 [27] PAVANI S R P, PIESTUN R. High-efficiency rotating point spread functions[J]. Optics Express, 2008, 16(5): 3484-3489. doi: 10.1364/OE.16.003484 [28] PAVANI S R P, THOMPSON M A, BITEEN J S, et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(9): 2995-2999. doi: 10.1073/pnas.0900245106 [29] BADDELEY D, CANNELL M B, SOELLER C. Three-dimensional sub-100 nm super-resolution imaging of biological samples using a phase ramp in the objective pupil[J]. Nano Research, 2011, 4(6): 589-598. doi: 10.1007/s12274-011-0115-z [30] LEW M D, LEE S F, BADIEIROSTAMI M, et al. Corkscrew point spread function for far-field three-dimensional nanoscale localization of pointlike objects[J]. Optics Letters, 2011, 36(2): 202-204. doi: 10.1364/OL.36.000202 [31] SIVILOGLOU G A, BROKY J, DOGARIU A, et al. Observation of accelerating airy beams[J]. Physical Review Letters, 2007, 99(21): 213901. doi: 10.1103/PhysRevLett.99.213901 [32] SIVILOGLOU G A, CHRISTODOULIDES D N. Accelerating finite energy airy beams[J]. Optics Letters, 2007, 32(8): 979-981. doi: 10.1364/OL.32.000979 [33] JIA SH, VAUGHAN J C, ZHUANG X W. Isotropic three-dimensional super-resolution imaging with a self-bending point spread function[J]. Nature Photonics, 2014, 8(4): 302-306. doi: 10.1038/nphoton.2014.13 [34] LIU SH, KROMANN E B, KRUEGER W D, et al. Three dimensional single molecule localization using a phase retrieved pupil function[J]. Optics Express, 2013, 21(24): 29462-29487. doi: 10.1364/OE.21.029462 [35] ZELGER P, KASER K, ROSSBOTH B, et al. Three-dimensional localization microscopy using deep learning[J]. Optics Express, 2018, 26(25): 33166-33179. doi: 10.1364/OE.26.033166 [36] SHECHTMAN Y, SAHL S J, BACKER A S, et al. Optimal point spread function design for 3D imaging[J]. Physical Review Letters, 2014, 113(13): 133902. doi: 10.1103/PhysRevLett.113.133902 [37] SHECHTMAN Y, WEISS L E, BACKER A S, et al. Precise three-dimensional scan-free multiple-particle tracking over large axial ranges with tetrapod point spread functions[J]. Nano Letters, 2015, 15(6): 4194-4199. doi: 10.1021/acs.nanolett.5b01396 [38] GORDON-SOFFER R, WEISS L E, ESHEL R, et al. Microscopic scan-free surface profiling over extended axial ranges by point-spread-function engineering[J]. Science Advances, 2020, 6(44): eabc0332. doi: 10.1126/sciadv.abc0332 [39] ZHOU Y ZH, CARLES G. Precise 3D particle localization over large axial ranges using secondary astigmatism[J]. Optics Letters, 2020, 45(8): 2466-2469. doi: 10.1364/OL.388695 [40] WEISS L E, SHALEV EZRA Y, GOLDBERG S, et al. Three-dimensional localization microscopy in live flowing cells[J]. Nature Nanotechnology, 2020, 15(6): 500-506. doi: 10.1038/s41565-020-0662-0 [41] JIN D Y, XI P, WANG B M, et al. Nanoparticles for super-resolution microscopy and single-molecule tracking[J]. Nature Methods, 2018, 15(6): 415-423. doi: 10.1038/s41592-018-0012-4 [42] NEHME E, WEISS L E, MICHAELI T, et al. Deep-STORM: super-resolution single-molecule microscopy by deep learning[J]. Optica, 2018, 5(4): 458-464. doi: 10.1364/OPTICA.5.000458 [43] NEHME E, FREEDMAN D, GORDON R, et al. DeepSTORM3D: dense 3D localization microscopy and PSF design by deep learning[J]. Nature Methods, 2020, 17(7): 734-740. doi: 10.1038/s41592-020-0853-5 [44] BALZAROTTI F, EILERS Y, GWOSCH K C, et al. Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes[J]. Science, 2017, 355(6325): 606-612. doi: 10.1126/science.aak9913 [45] GWOSCH K C, PAPE J K, BALZAROTTI F, et al. MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells[J]. Nature Methods, 2020, 17(2): 217-224. doi: 10.1038/s41592-019-0688-0 [46] TESTA I, WURM C A, MEDDA R, et al. Multicolor fluorescence nanoscopy in fixed and living cells by exciting conventional fluorophores with a single wavelength[J]. Biophysical Journal, 2010, 99(8): 2686-2694. doi: 10.1016/j.bpj.2010.08.012 [47] BROEKEN J, RIEGER B, STALLINGA S. Simultaneous measurement of position and color of single fluorescent emitters using diffractive optics[J]. Optics Letters, 2014, 39(11): 3352-3355. doi: 10.1364/OL.39.003352 [48] SMITH C, HUISMAN M, SIEMONS M, et al. Simultaneous measurement of emission color and 3D position of single molecules[J]. Optics Express, 2016, 24(5): 4996-5013. doi: 10.1364/OE.24.004996 [49] ZHANG ZH Y, KENNY S J, HAUSER M, et al. Ultrahigh-throughput single-molecule spectroscopy and spectrally resolved super-resolution microscopy[J]. Nature Methods, 2015, 12(10): 935-938. doi: 10.1038/nmeth.3528 [50] SHECHTMAN Y, WEISS L E, BACKER A S, et al. Multicolour localization microscopy by point-spread-function engineering[J]. Nature Photonics, 2016, 10(9): 590-594. doi: 10.1038/nphoton.2016.137 [51] HERSHKO E, WEISS L E, MICHAELI T, et al. Multicolor localization microscopy and point-spread-function engineering by deep learning[J]. Optics Express, 2019, 27(5): 6158-6183. doi: 10.1364/OE.27.006158 [52] KIM T, MOON S, XU K. Information-rich localization microscopy through machine learning[J]. Nature Communications, 2019, 10(1): 1996. doi: 10.1038/s41467-019-10036-z [53] LEW M D, BACKLUND M P, MOERNER W E. Rotational mobility of single molecules affects localization accuracy in super-resolution fluorescence microscopy[J]. Nano Letters, 2013, 13(9): 3967-3972. doi: 10.1021/nl304359p [54] ENGELHARDT J, KELLER J, HOYER P, et al. Molecular orientation affects localization accuracy in superresolution far-field fluorescence microscopy[J]. Nano Letters, 2011, 11(1): 209-213. doi: 10.1021/nl103472b [55] ZHANGHAO K, CHEN L, YANG X S, et al. Super-resolution dipole orientation mapping via polarization demodulation[J]. Light:Science &Applications, 2016, 5(10): e16166. [56] ZHANGHAO K, GAO J T, JIN D Y, et al. Super-resolution fluorescence polarization microscopy[J]. Journal of Innovative Optical Health Sciences, 2018, 11(1): 1730002. doi: 10.1142/S1793545817300026 [57] ZHANGHAO K, CHEN X Y, LIU W H, et al. Super-resolution imaging of fluorescent dipoles via polarized structured illumination microscopy[J]. Nature Communications, 2019, 10(1): 4694. doi: 10.1038/s41467-019-12681-w [58] BACKLUND M P, LEW M D, BACKER A S, et al. Simultaneous, accurate measurement of the 3D position and orientation of single molecules[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(47): 19087-19092. doi: 10.1073/pnas.1216687109 [59] WILLIG K I, RIZZOLI S O, WESTPHAL V, et al. STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis[J]. Nature, 2006, 440(7086): 935-939. doi: 10.1038/nature04592 [60] HULLEMAN C N, THORSEN R Ø, KIM E, et al. Simultaneous orientation and 3D localization microscopy with a Vortex point spread function[J]. Nature Communications, 2021, 12(1): 5934. doi: 10.1038/s41467-021-26228-5 [61] HELL S, STELZER E H K. Properties of a 4Pi confocal fluorescence microscope[J]. Journal of the Optical Society of America A, 1992, 9(12): 2159-2166. doi: 10.1364/JOSAA.9.002159 [62] HAO X, LI Y M, FU SH, et al. Review of 4Pi fluorescence nanoscopy[J]. Engineering, 2022, 11: 146-153. doi: 10.1016/j.eng.2020.07.028 [63] AQUINO D, SCHÖNLE A, GEISLER C, et al. Two-color nanoscopy of three-dimensional volumes by 4Pi detection of stochastically switched fluorophores[J]. Nature Methods, 2011, 8(4): 353-359. doi: 10.1038/nmeth.1583 [64] HUANG F, SIRINAKIS G, ALLGEYER E S, et al. Ultra-high resolution 3D imaging of whole cells[J]. Cell, 2016, 166(4): 1028-1040. doi: 10.1016/j.cell.2016.06.016 [65] LI Y M, BUGLAKOVA E, ZHANG Y D, et al. Accurate 4Pi single-molecule localization using an experimental PSF model[J]. Optics Letters, 2020, 45(13): 3765-3768. doi: 10.1364/OL.397754 [66] CHEN J W, YAO B X, YANG ZH CH, et al. Ratiometric 4Pi single-molecule localization with optimal resolution and color assignment[J]. Optics Letters, 2022, 47(2): 325-328. doi: 10.1364/OL.446987 [67] ZHANG Y D, SCHROEDER L K, LESSARD M D, et al. Nanoscale subcellular architecture revealed by multicolor three-dimensional salvaged fluorescence imaging[J]. Nature Methods, 2020, 17(2): 225-231. doi: 10.1038/s41592-019-0676-4 [68] STETSON P B. DAOPHOT: A computer program for crowded-field stellar photometry[J]. Publications of the Astronomical Society of the Pacific, 1987, 99(613): 191. [69] LEUTENEGGER M, RAO R, LEITGEB R A, et al. Fast focus field calculations[J]. Optics Express, 2006, 14(23): 11277-11291. doi: 10.1364/OE.14.011277 [70] HANSER B M, GUSTAFSSON M G L, AGARD D A, et al. Phase retrieval for high-numerical-aperture optical systems[J]. Optics Letters, 2003, 28(10): 801-803. doi: 10.1364/OL.28.000801 [71] BABCOCK H P, ZHUANG X W. Analyzing single molecule localization microscopy data using cubic splines[J]. Scientific Reports, 2017, 7(1): 552. doi: 10.1038/s41598-017-00622-w [72] LI Y M, MUND M, HOESS P, et al. Real-time 3D single-molecule localization using experimental point spread functions[J]. Nature Methods, 2018, 15(5): 367-369. doi: 10.1038/nmeth.4661 [73] SPEISER A, MÜLLER L R, HOESS P, et al. Deep learning enables fast and dense single-molecule localization with high accuracy[J]. Nature Methods, 2021, 18(9): 1082-1090. doi: 10.1038/s41592-021-01236-x [74] THIELE J C, HELMERICH D A, OLEKSIIEVETS N, et al. Confocal fluorescence-lifetime single-molecule localization microscopy[J]. ACS Nano, 2020, 14(10): 14190-14200. doi: 10.1021/acsnano.0c07322 [75] LIN Y, SHARIFI F, ANDERSSON S B. Three-dimensional localization refinement and motion model parameter estimation for confined single particle tracking under low-light conditions[J]. Biomedical Optics Express, 2021, 12(9): 5793-5811. doi: 10.1364/BOE.432187 [76] LI Y M, WU Y L, HOESS P, et al. Depth-dependent PSF calibration and aberration correction for 3D single-molecule localization[J]. Biomedical Optics Express, 2019, 10(6): 2708-2718. doi: 10.1364/BOE.10.002708 [77] LI Y M, SHI W, LIU SH, et al. Global fitting for high-accuracy multi-channel single-molecule localization[J]. Nature Communications, 2022, 13(1): 3133. doi: 10.1038/s41467-022-30719-4 -





 下载:
下载: